Ehsan Hajiramezanali

AI Research Scientist
Genentech
I am an AI Research Scientist at Genentech in the Foundational ML Research team. I have received my Ph.D. on "Bayesian Learning with Heterogeneous Data" at Texas A&M University under supervision of Prof. Xiaoning Qian. My research lies at the intersection of machine learning and Bayesian statistics. My main area of interest is learning with heterogeneous and structured data and Bayesian interpretation, especially in computational biology. This includes topics in causal learning, relational learning, multi-domain learning, generative models, representation learning, and time-series analysis.
Highlights
- 10/2022: Our recent paper on STab: Self-supervised Learning for Tabular Data is accepted at NeurIPS 2022 Workshop on Table Representation Learning.
- 01/2022: Our recent paper on Multi-omics Relational Learning is accepted at ICLR 2022.
- 10/2021: Our paper on Self-Supervised Representation Learning with Tabular Data is accepted at NeurIPS 2021. The implementation of this work is available here.
- 07/2021: I have received best reviewer award in ICML 2021.
- 11/2020: I have defended my Ph.D. thesis.
- 10/2020: Delighted to announce that I will be joining AstraZeneca as an AI Research Scientist in Respiratory and Immunology.
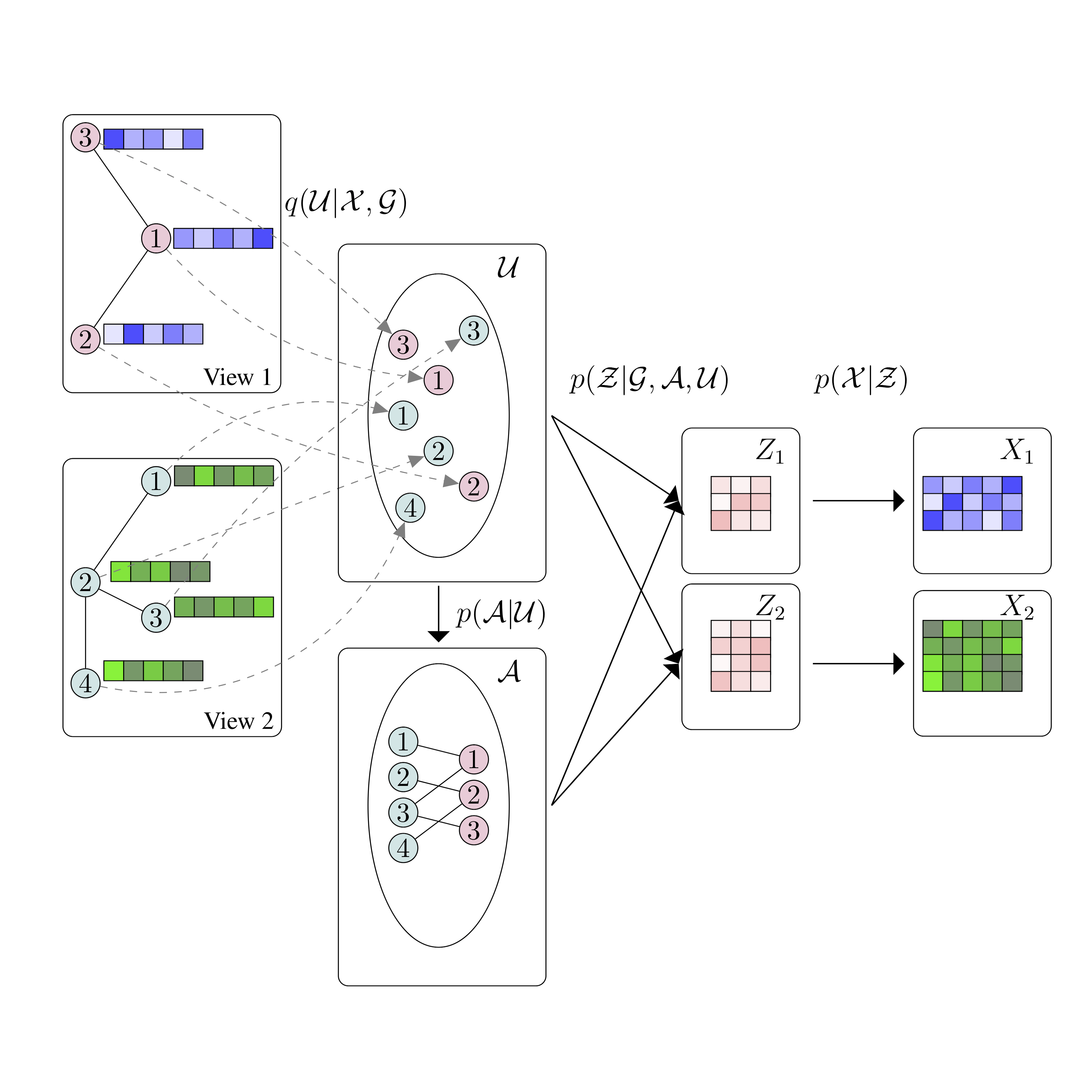
- 09/2020: Our paper, "BayReL: Bayesian Relational Learning", has been accepted in NeurIPS 2020. BayReL is the first (Bayesian) model for learning (inter-)relations between nodes in a set of graphs. Congrats and thanks to Arman, and profs. Qian, Narayanan, and Duffield.
- 07/2020: I am honored to receive Chevron Scholarship! Thanks to my advisor, Prof. Xiaoning Qian, for his supports.
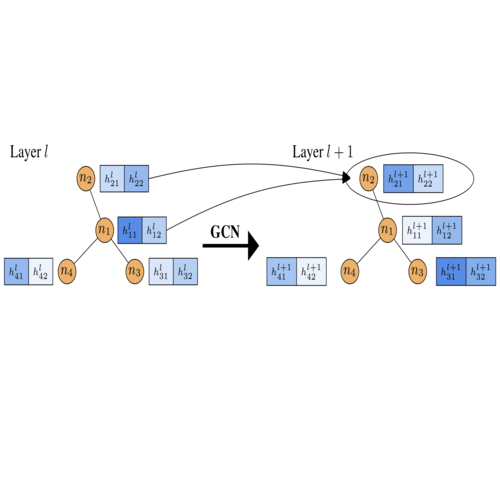
- 05/2020: "Bayesian Graph Neural Networks with Adaptive Connection Sampling", co-authored with Arman Hasanzadeh, Nick Duffield, Krishna Narayanan, Mingyuan Zhou, and Xiaoning Qian, is accepted in ICML 2020.
- 04/2020: Our paper has been selected as finalist for the ICASSP 2020 Best Student Paper Award.
- 03/2020: I will join the Tagare Lab in the School of Medicine at Yale University as a Visitor Scholar in summer 2020.
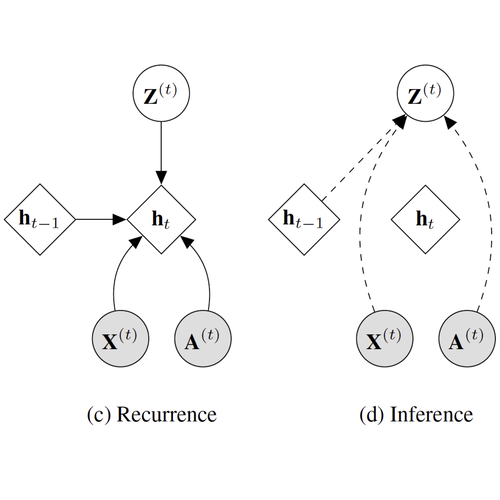
- 01/2020: "Semi-Implicit Stochastic Recurrent Neural Networks", co-authored with Arman Hasanzadeh, Nick Duffield, Krishna Narayanan, Mingyuan Zhou, and Xiaoning Qian, is accepted for Oral Presentation in ICASSP 2020.
- 01/2020: The book "ESET 211 AC Circuits - Lab Manual," co-authored with A. Bal, Y. Chen, Z. Chen, A. Divanahi, E. Kaya, S. Moosavi, and P. Wallace is published by Industrial Distribution Program, Texas A&M University.
- 10/2019: I am honored to receive Outstanding Graduate Student Award in ECE @ Texas A&M University!
- 09/2019: We are presenting two papers in NeurIPS 2019.